Single-Cell Genomics and Metabolic Modelling for tailored microbial consortia design
Genomics and metabolic modeling for the tailored design of microbial consortia aims to uncover the metabolic potential of non-culturable microorganisms and their interactions within complex communities. SIGMA seeks to adapt advanced bulk and single-cell sequencing approaches to investigate bacteria, archaea, and phages involved in CO2 capture processes, all within the broader context of the anaerobic digestion (AD) microbiome. The ultimate goal is to explore high-value applications of the AD microbiome and to develop the necessary techniques for creating highly productive and specialized consortia.

SIGMA was funded by UNIPD STARS Grant, an initiative by the University of Padua (UNIPD) aimed at supporting early-career researchers and promoting innovative projects. It offers funding to researchers who wish to carry out high-impact scientific work and enhancing their research capabilities.
Our Dream Team

PI: I specialized in molecular biology and microbial ecology to investigate carbon cycling, bioenergy production, and CO2 capture. As Senior Researcher (RtdB) at the Department of Biology, I’ve advanced bioinformatics for omics, exploring single microorganisms and communities. I study molecular and metabolic mechanisms shaping environmental microbiomes, building metabolic models for process optimization and integrating omics with flux balance analysis.

Research Assistant: I completed my Bachelor’s degree in Biotechnology and Master’s degree in Industrial Biotechnology, with a thesis focused on the production of polyhydroxyalkanoates by Cupriavidus necator in response to physiological stress for CO2 capture. Currently, I am involved in a project titled “Microbial Culturomics and High-Yield Single-Cell Technologies for the Investigation of the Anaerobic Microbiome,” where I use single-cell methods to support genomic and transcriptomic studies at the individual organism level within mixed microbial communities.
Further collaborators include:



Research achievements
Studying the microbiome of biogas upgrading and CO2 fixation is key to achieving more sustainable energy production. Efficient microbial conversion of CO2 into organic compounds relies on effective microbial resource management, which requires an in-depth understanding of metabolic pathways, mechanisms driving syntrophic interactions, and the relationships between phages and microbial hosts in anaerobic microbiomes. Traditional metagenomic approaches have been widely used to elucidate these processes but often fail to capture fine-scale variations in complex microbial communities.
To overcome these limitations, the SIGMA project focused on adapting and applying single-cell sequencing (scDNA-seq) and culturomics approaches to analyse the structure of anaerobic microbiomes adapted to CO2 as its primary carbon source.
The scDNA-seq workflow involved encapsulating cells in semi-permeable capsules (SPCs), followed by lysis, DNA amplification, and combinatorial barcoding prior to sequencing with Illumina technology (Fig. 1).
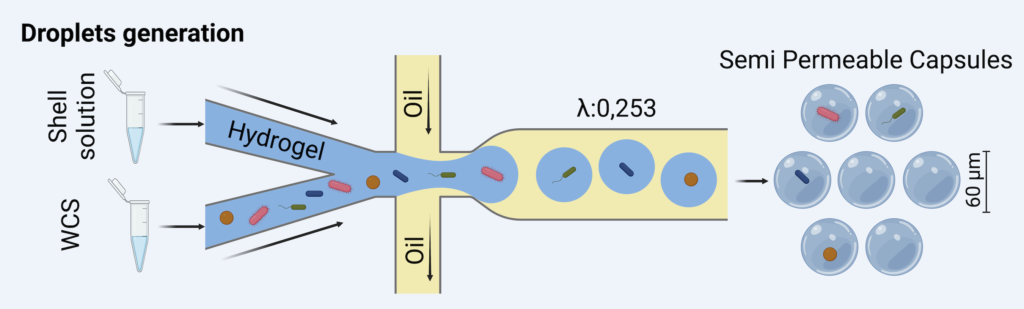
Fig. 1. Single-cell sequencing workflow. Cells are encapsulated into semi-permeable capsules and lysed using chemical-enzymatic agents. The resulting DNA is amplified and tagged through combinatorial barcoding. The pooled DNA is then sequenced using Illumina technology, yielding single amplified genomes (SAGs).
In the context of SIGMA project, lysis protocols were refined by combining enzymatic and chemical treatments to maximize DNA release.

Since this approach had never been applied to strictly anaerobic cultures dominated by methanogenic archaea, scDNA-seq was complemented by bulk metagenomics as a gold standard. The validation of reconstructed single amplified genomes (SAGs) was achieved by comparing their quality and abundance with metagenome-assembled genomes (MAGs) of the same taxa.
First case study
Metagenomic sequencing of a simplified CO2-fixating microbiome identified 18 bacterial species with a relative abundance above 0.1%. Methanothermobacter thermautotrophicus emerged as the dominant hydrogenotrophic archaeon. The scDNA-seq generated 706 SAGs, with a mean completeness of 11.9% and contamination of 0.26%, and demonstrating a strong linear correlation between completeness and coverage (P-value < 4.7e-81). Although individual SAGs exhibited a low Breadth-Expected Breadth Ratio (BER) due to the inherent limitations of whole genome amplification, it was demonstrated that combining multiple SAGs from the same species allows near-complete genome reconstruction (Fig. 2).
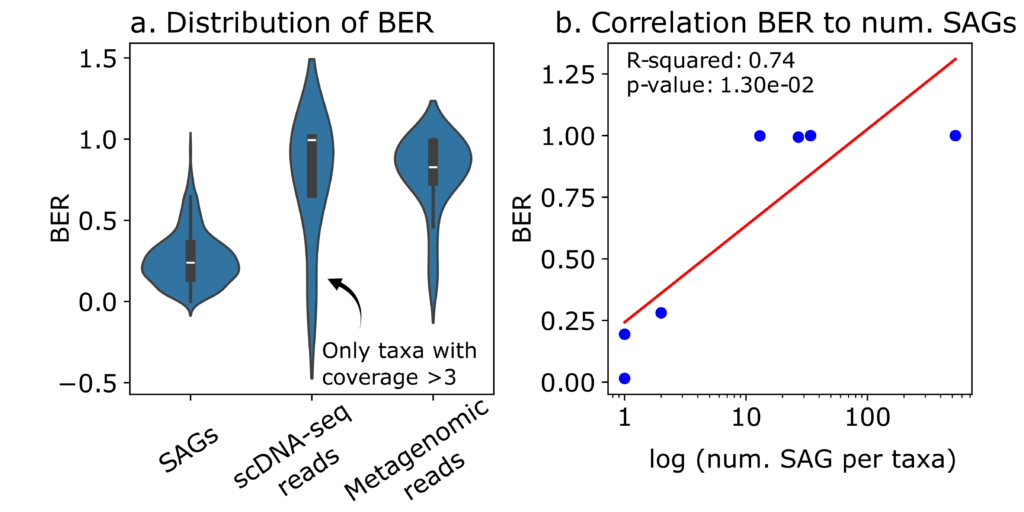
In total, 86% of SAGs matched existing MAGs, and 73 SAGs belonged to species not detected through metagenomics. However, comparisons between metagenomics and scDNA-seq revealed significant differences in estimating species relative abundance, with only 7 out of 19 species concordantly identified by both methods (Fig. 3). This discrepancy suggests that scDNA-seq underrepresents less abundant community members, likely due to the dominance of M. thermautotrophicus (RA >93%), and indicates that future trials should target a higher number of cells.

This primary investigation underscores the potential of scDNA-seq to enhance microbiome profiling beyond traditional genome-centric approaches and has contributed to the development of a bioinformatics toolbox for analyzing scDNA datasets available on GitHub. These preliminary results were presented at the ISME19 conference, reinforcing the study’s visibility and significance in microbial ecology.
Second case study
Following the validation of the approach, focus shifted toward leveraging the insights provided by scDNA-seq to investigate bacterial and archaeal viruses and their hosts. Traditional metagenomic methods often overlook key genetic elements (such as plasmids and phages) that are critical for microbial metabolism and interspecies interactions. To address this, samples from the first case study was used to study the interaction of phages with the microbiome and free-living phages, focusing on microbe’s defense mechanisms, such as CRISPR-Cas, and how phages infect them. Four stress conditions known to induce temperate phages were used: UV exposure, Mitomycin C, heavy metal (CuCl2), and hydrogen peroxide (H2O2). These treatments were selected to identify the optimal conditions for triggering phage induction and controlling microbiota composition, particularly for the removal of unwanted bacteria. Unlike the previous experiment, a higher number of cells per sample were targeted during scDNA-seq to increase statistical resolution and facilitate the development of a method for reconstructing strain-level genomes by pooling sequences from individual cells. Metagenomics identified 24 MAGs with a relative abundance (RA) above 0.01%, with both methods confirming the dominance of M. thermautotrophicus. Notably, scDNA-seq recovered Acetivibrionales sp.21, a species detected only in the supernatant of metagenomic samples, demonstrating its capacity to capture overlooked MAGs.
Regarding host-phage associations, metagenomic analysis identified 98 temperate and 30 virulent viral MAGs (virMAGs). Using CRISPR-spacer and amino acid identity matches, phage-host associations were predicted, revealing infection hubs such as Symbiobacterium, which appeared to be infected by 15 different phages. scDNA-seq confirmed 2 of 4 unique associations inferred from CRISPR-spacer analysis and suggested potential hosts for an additional 39 phages. Overall, this study highlights the complementary strengths of scDNA-seq and metagenomics in deciphering the relationships within CO2-fixing microbiomes. While metagenomics offers broader community and phage coverage, scDNA-seq provides unique insights into the genetic variability of mobile elements.
Future Directions. Together, these approaches enhance our understanding of microbial communities and their biotechnological potential. Given the challenges encountered, future research will focus on optimizing cell quantification prior to encapsulation, testing alternative methods for whole genome amplification (such as primary template-directed amplification) and refining bioinformatics pipelines to improve species and strain-level genome recovery accuracy.
Dissemination success
Conferences:
19th International Symposium on Microbial Ecology (ISME19), August 18th-23th 2024, Cape Town, South Africa. INTERNATIONAL CONGRESS, POSTER: De Bernardini N, Giangieri G, Savio F, Ling M, Fraulini S, Orellana E, Ji M, Campanaro S, Treu L. Genetic variability in a carbon dioxide reducing microbial community: a comparison between metagenomics and single-cell sequencing.
The 5th International Conference on Biogas Microbiology (ICBM-5), May 26th-29th 2025, Galway, Ireland. INTERNATIONAL CONGRESS, ORAL: Savio F, De Bernardini N, Orellana E, Ji M, Campanaro S, Treu L. Integrating single-cell DNA sequencing and metagenomics to uncover overlooked genetic features in a CO₂-fixing microbiome for biogas upgrading.
Students that published their thesis on the topic:
- Maria Rossi, graduate student (master). Topic: “Microbial communities: a
biotechnology study with single-cell sequencing” - Retno Wahidah Supardi, graduate student (master). Topic: “Exploring the Role of Phages in the Anaerobic Digestion Microbial Community through Metagenomics and Single-Cell Analysis”
- Alessandro Peretti, graduate student (bachelor). Topic: “Esplorazione delle Frontiere: Ottimizzazione Single-Cell nell’Analisi delle Comunità Microbiche”
An additional related event organized by Prof. Treu was the LIFE CO2toCH4 annual meeting, held on 27th November 2024 at the Botanical Garden in Padova. During this meeting, Prof. Treu delivered a seminar titled “Process optimization using technically advanced systems and control architectures based on microbial resource management “, aimed at promoting the dissemination of results and exploring collaboration opportunities. Furthermore, two internal seminars were conducted within the Department of Biology, Genomic and Bioinformatic Research Unit: Dr. Filippo Savio presented on “Single-cell RNA sequencing: a brief overview ” while Dr. Nicola De Bernardini discussed “The coverage constraints in single-cell sequencing analysis “.

Funding organization: University of Padova
Call: Supporting TAlent in ReSearch@University of Padua STARS@UNIPD
Timeline: 2024-2025 – 30 months
Designed with WordPress